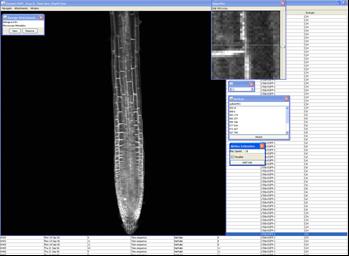
This software is designed to estimate the motion of patches of pixels obtained in fluorescence images from a Confocal Laser Microscope. Displacements are reported to 0.1 pixel resolution, and the confidence of these estimates is also reported.
PlantVis generates large output files containing the coordinates of tracked pixels, the estimated displacement of these pixels between images, and an estimate of the confidence that these displacements are correct. This output needs to be analysed further – we do this using a series of scripts written in R. This is a very flexible approach, but does require the user to obtain a reasonable degree of proficiency with using R.
Examples of output
- velocity and strain rate as a function of distance from the quiescent centre of the root, for only pixels tracked with a high degree of confidence;
- velocity in particular root tissues, [as highlighted in image];
For more information, please contact Stephen McKenna (algorithms and coding), Glyn Bengough (biophysical applications) or Tracy Valentine (microscopy and applications).