We have developed prototype software for cell segmentation and tracking of dual labelled Arabidopsis roots (nuclear and plasma-membrane markers) in 4-D.
Nuclei are tracked located and tracked in 3-D versus time. Cell vertices are identified and used to reconstruct cell networks, which are then tracked with time. Software run from ImageJ.
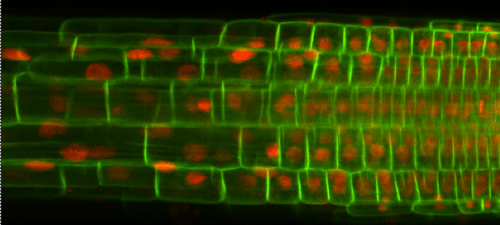
Figure: Arabidopsis root dual-labeled with nuclear and cell membrane markers
For more information, please contact Stephen McKenna (algorithms and coding), Glyn Bengough (biophysical applications), Tracy Valentine (microscopy and applications), Jim Haseloff (plant material and cell biology).