CellModeller is a python module for 2D multicellular analysis and computing. It has a generic data structure (multiscale graph representation) and uses elementary operations to query and modify these objects (for example, get the neighbour of a cell or modify the concentration of its constituents). The behaviour and properties of each entity in the structure (vertex, wall, cell...) can be controlled through user specified models, using python scripting language.
Application example: simulating branch outgrowth
In the shoot apical meristem, primordium initiation is accompanied by cell proliferation under the L1 layer. Expanding cells largely adhere to surrounding tissues, and the mechanical behaviour of all tissues influences the kinematics of expansion in the emerging meristem. The meristem may develop a localised bulge if localised pressure or wall softening in epidermal cells initiates the primordium (Selker et al., 1992). This was modelled by a simple architecture of cells consisted of three different tissues: the first tissue was a single layer of cells representing the L1 layer. The second tissue was three layers of cells under the L1 layer (orange). At the start of the simulation, four cells of the second tissue enter a proliferation stage (third tissue in green). The wall properties (viscosities and moment of inertia) are set at the level of the tissue:
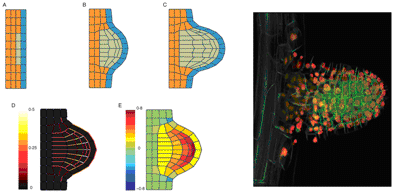
Figure: different types of cells organised in tissues can generate complex patterns of stress and deformation in tissues. Here, a group of internal cells have a higher expansion rate, and induce the formation of an outgrowing branch.
Cell modeller has its own website with documentation.